 Image credit: [Lorenzo Madeddu]
Image credit: [Lorenzo Madeddu]
Abstract
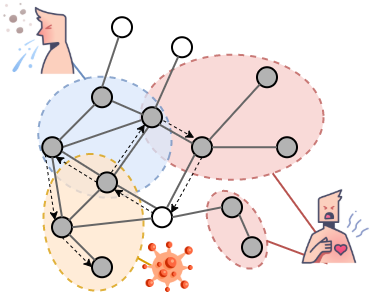
(Testo in lingua originale) We predict disease-genes relations on the human interactome network using a methodology that jointly learns functional and connectivity patterns surrounding proteins. Contrary to other data structures, the interactome is characterised by high incompleteness and absence of explicit negative knowledge, which makes predictive tasks particularly challenging. To exploit at best latent information in the network, we propose an extended version of random walks, named Random Watcher-Walker (RW²), which is shown to perform better than other state-of-the-art algorithms. We also show that the performance of RW² and other compared state-of-the-art algorithms is extremely sensitive to the interactome used, and to the adopted disease categorisations, since this influences the ability to capture regularities in presence of sparsity and incompleteness.